Schistosoma mansoni lncRNAs Database
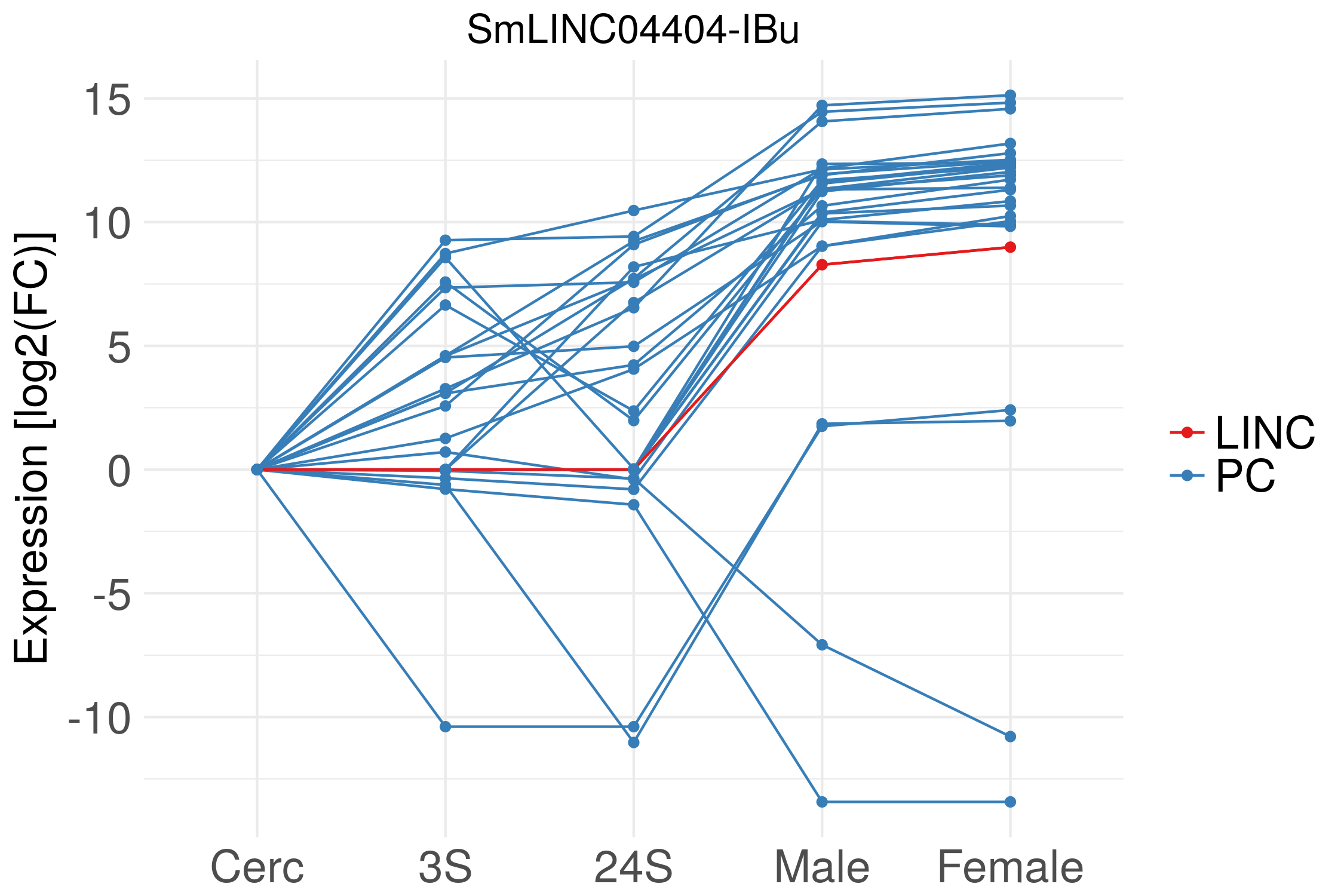
SmLINC04404-IBu Data
Chromosome: Chr 7 - Coordinate: Chr 7:7903003-7903815 - Strand: +

Expression Fold Change Data [log2(FC)] - Download data
Pearson Correlation Coefficient Data - Download data

Expression Fold Change Data [log2(FC)] - Download data
Pearson Correlation Coefficient Data - Download data